what to do once you discover a binding partner protein
Abstract
Predicting the construction of interacting protein chains is a key step towards agreement protein part. Unfortunately, no computational method can produce accurate structures of poly peptide complexes. AlphaFold2, has shown unprecedented levels of accuracy in modelling single chain protein structures. Hither, we apply AlphaFold2 for the prediction of heterodimeric poly peptide complexes. We find that the AlphaFold2 protocol together with optimised multiple sequence alignments, generate models with acceptable quality (DockQ ≥ 0.23) for 63% of the dimers. From the predicted interfaces we create a unproblematic function to predict the DockQ score which distinguishes acceptable from wrong models too as interacting from non-interacting proteins with state-of-art accuracy. Nosotros find that, using the predicted DockQ scores, we can place 51% of all interacting pairs at 1% FPR.
Introduction
Protein–poly peptide interactions are key mediators in biological processes. Most interactions are governed by the iii-dimensional organization and the dynamics of the interacting proteins1. Such interactions vary from beingness permanent to transient2,3. Some protein–poly peptide interactions are specific for a pair of proteins, while some proteins are promiscuous and interact with many partners. This complication of interactions is a challenge both for experimental and computational methods.
Oft, studies of protein–protein interactions can be divided into ii categories, the identification of what proteins collaborate and the identification of how they interact. Although these issues are distinguished, some methods have been applied to both problems4,v. Protein docking methodologies refer to how proteins collaborate and can exist divided into two categories considering proteins as rigid bodies; those based on an exhaustive search of the docking space6 and those based on alignments (both sequence and structure) to structural templates7. Exhaustive approaches rely on generating all possible configurations betwixt poly peptide structures or models of the monomers8,ix and selecting the right docking through a scoring function, while template-based docking merely needs suitable templates to identify a few probable candidates. Withal, flexibility has often to be considered in protein docking to business relationship for interaction-induced structural rearrangements10,eleven. Therefore, flexibility limits the accuracy achievable past rigid-body docking12, and flexible docking is traditionally too boring for large-calibration applications. A possible compromise is represented by semi-flexible docking approachesxiii that are more than computationally viable and can consider flexibility to some degree during docking.
Regardless of unlike strategies, docking remains a challenging problem. In the CASP13-CAPRI experiments, human grouping predictors achieved up to fifty% success rate (SR) for height-ranked docking solutions14. Alternatively, a recent benchmark report8 reports SRs of different web-servers reaching up to 16% on the well-known Benchmark five datasetxv.
Recently, in the CASP14 experiment, AlphaFold2 (AF2) reached an unprecedented operation level in structure prediction of single-concatenation proteins16. Cheers to an advanced deep learning model that efficiently utilises evolutionary and structural information, this method consistently outperformed all competitors, reaching an average GDT_TS score of 9016. Recently, RoseTTAFold was developed, trying to implement similar principles17. Since then, other end-to-finish structure predictors have emerged using different principles such as fast multiple sequence alignment (MSA) processing in DMPFold2xviii and linguistic communication model representationsnineteen.
As an culling to other docking methods, it is possible to utilise co-evolution to predict the interaction between ii protein chains. Initially, directly coupling analysis (DCA) was used to predict the interaction of bacterial ii-component signalling proteins20,21. Afterward, these methods were improved using motorcar learning22.
In a Fold and Dock approach, two proteins are folded and docked simultaneously. Nosotros recently adult a Fold and Dock pipeline using another distance prediction method focused on poly peptide folding (trRosetta23). In this pipeline, the interaction between two chains from a heterodimeric protein complex and their structures were predicted using altitude and angle constraints from trRosetta24,25. This written report demonstrated that a pipeline focused on intra-concatenation structural feature extraction tin be successfully extended to derive inter-concatenation features likewise. All the same, just seven% of the tested proteins were successfully folded and docked.
In that written report, we institute that generating the optimal MSA is crucial for obtaining accurate Fold and Dock solutions, but this is not e'er trivial due to the necessity to identify the verbal set of interacting protein pairs26. Given the existence of multiple paralogs for nearly eukaryotic proteins, this is difficult. Nosotros too establish that this process requires an optimal MSA depth to optimise inter-concatenation data extraction. Too deep MSAs might contain false positives (i.e. protein pairs that collaborate differently), resulting in noise masking the sought after co-evolutionary signal, while also shallow alignments practise non provide sufficient co-evolutionary signals.
In this piece of work, we systematically employ the AF2 pipeline on two unlike datasets to Fold and Dock protein–protein pairs simultaneously. Nosotros explore the docking success using the AF2 pipeline in combination with unlike input MSAs, in society to study the relationship between the output model quality and these inputs. Nosotros also find that, by scoring multiple models of the same protein–protein interaction with a predicted DockQ score (pDockQ), we tin can distinguish with high confidence acceptable (DockQ ≥ 0.23) from incorrect models. The modelling success is college for bacterial protein pairs, pairs with large interaction areas consisting of helices or sheets, and many homologous sequences. We besides examination the possibility to distinguish interacting from not-interacting proteins and find that, using pDockQ, we tin can carve up truly interacting from non-interacting proteins with consistent accurateness. We find that the results in terms of successful docking using AF2 are superior to other docking methods. AF2 conspicuously outperforms a recent country-of-the-fine art method27 and our protocol performs quite close to (63% vs 72%) the recently developed AF-multimer28, which was developed using the same information every bit the test set here, making a direct comparison difficult.
Results and discussion
Identifying the all-time AlphaFold2 model
The SR, i.e., the per centum of adequate models (DockQ > 0.23), is used to mensurate AF2 functioning over the development set up (216 proteins) using the unlike MSAs. The all-time operation is 33.3% for the AF2 MSAs and 39.4% for the AF2+ paired MSAs (Tabular array 1). It is thereby evident that combining both paired and AF2 MSAs is superior to using them separately. The average performance of the AF2 and the paired MSAs is similar, just for individual protein pairs, frequently one of the two MSAs is superior to the other, as seen from that the Pearson correlation coefficient for the DockQ scores betwixt AF2 vs paired MSAs is 0.54 (Supplementary Tabular array ane). Therefore, combining AF2 and paired MSAs improves the results.
Adjacent, nosotros compared the default AF2 model (model_1) with the fine-tuned versions of (model_1_ptm). Surprisingly, the original AF2 model_1 outperforms AF2 model_1_ptm in nigh cases (Tabular array 1). Further, the difference betwixt 10 recycles-i ensemble and 3 recycles-eight ensembles is minor across all MSAs and AF2 models. Therefore, the input information and the AF2 model appear to impact the outcome the well-nigh.
Test prepare functioning
The best model and configuration for AF2 (m1-10-1) was used for further studies on the test gear up. The best outcome using this modelling strategy results in an SR of 57.8% (856 out of 1481 correctly modelled complexes) for the AF2 + paired MSAs compared with 45.0% using the AF2 MSAs solitary (Fig. 1, Table 2). The results using the block diagonalization+paired MSAs are about identical (SR = 58.4%, median = 0.363). Further, running 5 initialisations with random seeds and ranking the models using the predicted DockQ score (pDockQ, Fig. 2c), increases the SR to 61.7% and 62.seven% for the AF2 + paired and block diagonalization+paired MSAs, respectively (model variation and ranking, Fig. 2). Using the combination of AF2 and paired MSAs increases functioning, suggesting that AF2 gains both from larger and paired MSAs, although it often can manage with less information.
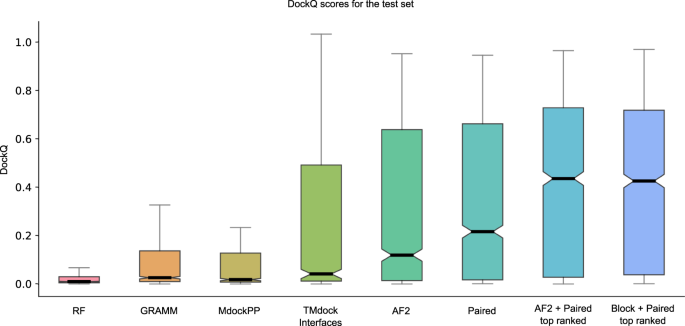
Distribution of DockQ scores every bit boxplots for different modelling strategies on the test set. Boxes comprehend data quartiles, horizontal lines mark the medians and upper and lower whiskers indicate respectively maximum and minimum values for each distribution. All AF2 models have been run with the same neural network configuration (m1-10-1). Outlier points are not displayed here. AF2, refers to running AF2 using the default AF2 MSAs, "Paired" refers to using MSAs paired using information about species and "Block" refers to using block diagonalization MSAs.
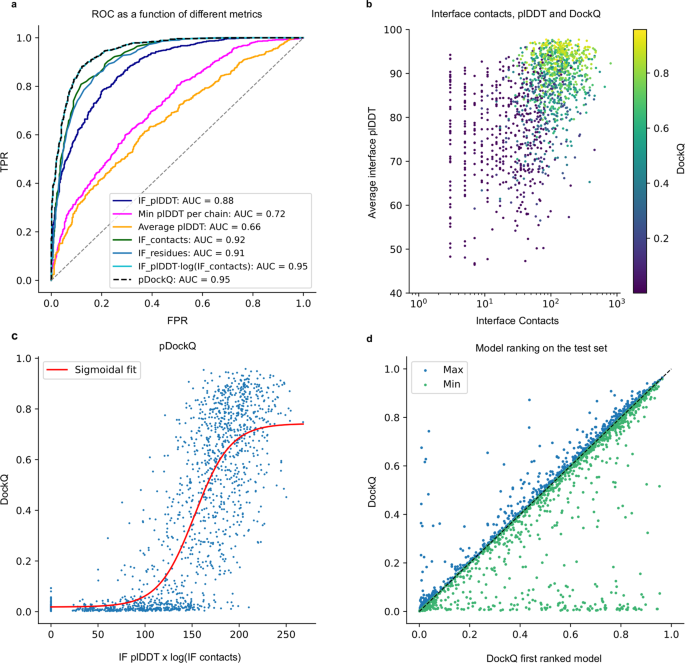
a ROC curve every bit a function of different metrics for the examination dataset (n = 1481, first run). Cβs within 8 Å from each other from different bondage are used to define the interface. IF_plDDT is the boilerplate plDDT of interface residues, min plDDT per chain is the minimum average plDDT of both chains, boilerplate plDDT is the average of the entire complex and IF_contacts and IF_residues are the number of interface residues and contacts respectively. pDockQ is a sigmoidal fit to the combined metric IF_plDDT⋅log(IF_contacts) fitted to predict DockQ as the target score, see C. b Average interface plDDT vs the logarithm of the interface contacts coloured past DockQ score on the test set (n = 1481). Increasing both the number of interface contacts and average interface plDDT results in higher DockQ scores. c Using the combined metric IF_plDDT⋅log(IF_contacts), we fit a sigmoidal curve towards the DockQ scores on the test fix (northward = 1481), enabling predicting the DockQ score in a continuous manner (pDockQ). The average error overall is 0.fourteen DockQ score. d Affect of unlike initialisations on the modelling outcome in terms of DockQ score on the test dataset (northward = 1481). The maximal and minimal scores are plotted confronting the top-ranked models using the pDockQ scores for the AF2 + paired MSAs, m1-10-1.
What is about striking is that AF2 outperforms all other tested docking methods by a big margin (Fig. ane, Tabular array ii). RF is better than AF2 merely for 14 pairs in the test set, while GRAMM and template-based docking (TMdock interface) outperform AF2 for 188 and 225 pairs, respectively. The best performing method in the CASP14-CAPRI experiment29, MDockPPthirty, achieves a SR of only 24.two%. The reason GRAMM, TMdock and MDockPP achieve this level of performance is likely due to the use of the leap form of the proteins, resulting in very high shape complementarity and therefore having the "answer" provided in a way.
The recently developed AF-multimer28 has the best performance (SR = 72.2%, median = 0.560, Tabular array two). This method was trained using the same data as the examination prepare, which makes a direct comparison difficult. Regardless, we practise believe it is likely that using AF-multimer, the performance would increase over the results of our pipeline, but it is possible the difference is less than the observed ix%.
Distinguishing acceptable from incorrect models
Information technology is not only essential to obtain improved predictions, simply besides to exist able to discriminate between acceptable and non-acceptable ones. We measure the separation between right (DockQ ≥ 0.23) and wrong models provided by several metrics using a receiver operating characteristic (ROC) curve. Different criteria were examined over the exam set, including (i) the number of unique interacting residues (Cβ atoms from unlike chains within 8 Å from each other) in the interface, (ii) the total number of interactions between Cβ atoms in the interface, (three) the average plDDT for the interface, (iv) the lowest plDDT of each single-concatenation average, and (five) the boilerplate plDDT over the whole protein heterodimer (Fig. 2a). Three criteria issue in very similar areas nether the curve (AUC) measures. The total number of interactions between Cβs and the number of residues in the interface can carve up the right/incorrect models with an AUC of 0.92 and 0.91 respectively, while the average interface plDDT results in an AUC of 0.88. Nonetheless, pLDDT results in college TPRs at lower FPRs; therefore, we multiply the plDDT with the logarithm of the interface contacts resulting in an AUC of 0.95.
Interestingly, the average plDDT of the entire complex only results in an AUC of 0.66, suggesting that both unmarried chains in a circuitous are oft predicted very well, while their relative orientation may still be wrong.
Effigy 2b shows that increasing both the number of interface contacts and the average interface plDDT results in higher DockQ scores for the test set. Using the combination of plDDT with the logarithm of the interface contacts, we, therefore, fit a simple sigmoidal function to the DockQ scores (Fig. 2c), encounter methods. This enables the prediction of the DockQ scores (pDockQ) in a continuous manner with an overall average error of 0.11 on the test set. The AUC using pDockQ equally a separator is identical to the combination of plDDT with the logarithm of the interface contacts, 0.95 (Fig. 2a).
Model variation and ranking for the test set up
Five models are generated using the all-time strategy (m1-10-1 with AF2 + paired MSAs) with different initialisation (random seeds). The boilerplate SR (57.ii% ± 0.0%) is similar for all five runs. However, the average deviation for private models is DockQ = 0.08 when comparing the best and worst models for a target (Fig. 2nd), i.e., in that location is some randomness to the success for an private pair. If the maximal DockQ score across all models is used, the SR would be 62.ix%. Although this is unachievable, ranking the models using the pDockQ score results in an SR of 61.7%. The AUC using the aforementioned metric for the ranked test set is 0.93, which means that 31% of all models are acceptable at an fault rate of 1% and 54% at an error rate of 10% (Supplementary Table 2).
Bacterial complexes are predicted more accurately
In the test set, almost 60% of the complexes tin be modelled correctly. We try to identify what distinguishes the successful and unsuccessful cases past analysing different subsets of the test fix. First, nosotros split the proteins by taxa, next past interface characteristics and finally by examining the alignments.
The SRs for each kingdom is; Eukarya 61%, Leaner 73.seven%, Archaea 84.5%, and Virus 60% (Supplementary Fig. 1b). Farther, the SRs for Saccharomyces cerevisiae is improve than for Homo sapiens (66% vs 58%, Fig. 3d). The college performance in prokaryotes is consistent with previous observations regarding the availability of evolutionary data in prokaryotes compared to Eukarya27 (Supplementary Fig. 1). The higher performance in S. cerevisiae compared to H. sapiens suggests a similar relationship between college and lower gild organisms within the same kingdom.
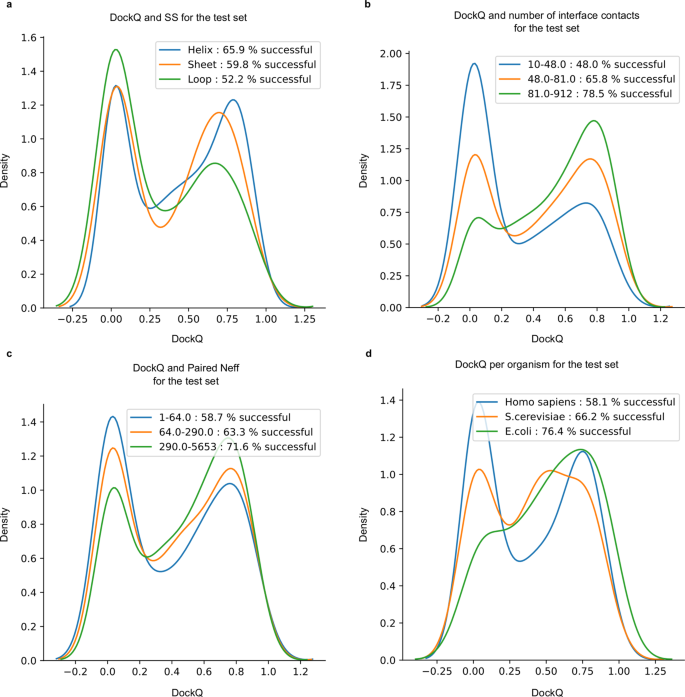
a Distribution of DockQ scores for three sets of interfaces with the majority of Helix, Canvass and Curl secondary structures. b Distribution of DockQ scores for tertiles derived from the distribution of contact counts in docking model interfaces. c Distribution of DockQ scores for tertiles derived from the distribution of Paired MSAs Neff scores. d Distribution of DockQ scores for the tiptop 3 organisms H. sapiens, Southward. cerevisiae and E. coli.
Side by side, we examine the interfaces. Different secondary structural content of the native interfaces is investigated (Fig. 3a). The highest SR is obtained mainly for helix interfaces (62%), followed by interfaces containing mainly sheets (59%). The loop interface SR of 53% is substantially lower than the others, suggesting that interfaces with more than flexible structures are harder to predict. We divide the dataset past interface size, and find that pairs with larger interfaces are easier to predict, as the SR increases from 47 to 74% between the smallest and biggest tertiles (Fig. 3b).
We continue to examine features of the MSAs. Kickoff, the impact of the number of non-redundant sequences (Neff) in both paired and AF2 MSAs was analysed. It is clear that the fraction of correctly modelled sequences increases with larger Neff scores (Fig. 3c). As well, paired MSA Neff (Fig. 3c) has a stronger influence on the outcome than the Neff of the AF2 MSAs (Supplementary Fig. 2a). Secondly, the MSA interface betoken in the paired MSAs, measured by the fraction of correct interface contacts using DCA, was analysed. MSAs with stronger interface signals show higher SRs, even if the paired MSAs are used in combination with the AF2 MSAs (Supplementary Fig. three). This suggests that MSA co-evolutionary point and, thereby, correct identification of orthologous protein sequences, has a strong impact on the effect.
CASP14 and novel proteins without templates
Bondage derived from CASP14 heteromeric targets and chains from PDB complexes with no templates are folded in pairs using the presented AF2 pipeline (default AF2 + paired MSAs, ten recycles, m1-ten-1 and five differently seeded runs).
For the CASP14 chains, four out of six pairs display a DockQ score larger than 0.23 (SR of 67%). No ranking is necessary in this example, given that all produced docking models for the same chain pair are very similar (the average standard difference is 0.01 betwixt each set of DockQ scores). An interesting unsuccessful docking is obtained modelling chains from the complex with PDB ID 6TMM (Supplementary Fig. 4), which are known to course a heterotetramer. In this structure, each concatenation A is in contact with its partner chain B at ii dissimilar sites. Both docking configurations (6TMM_A-B and 6TMM_A-D) put the concatenation in between the two binding sites. The other unsuccessful docking (6VN1_A-H) has an interface of only 19 residue pairs.
The SR for docking the proteins without templates is 50%. Between the five unlike initialisations, the boilerplate deviation in the DockQ score is 0.03, and in that location is no divergence in SR, i.e., ranking did non meliorate the SR. Two acceptable models are displayed in Fig. 5a (7EIV_A-C]) and B (7MEZ_A-B). More interesting, in ane of the incorrect models (7NJ0_A-C], Supplementary Fig. 5), the predictions go the location of both chains correct, merely their orientations wrong, resulting in DockQ scores close to 0. For 7EL1_A-E (Fig. 4c), the shorter chain E is not folded correctly, and instead of folding to a defined shape, it is stretched out and inserted inside chain A. It occupies the shape of the Dna in the native structure. In the two remaining incorrect models (7LF7_A-G and 7LF7_B-Chiliad), Fig. 4d, the chains only interact with a brusk loop of the Yard chain, making the docking very hard and possibly biologically meaningless.
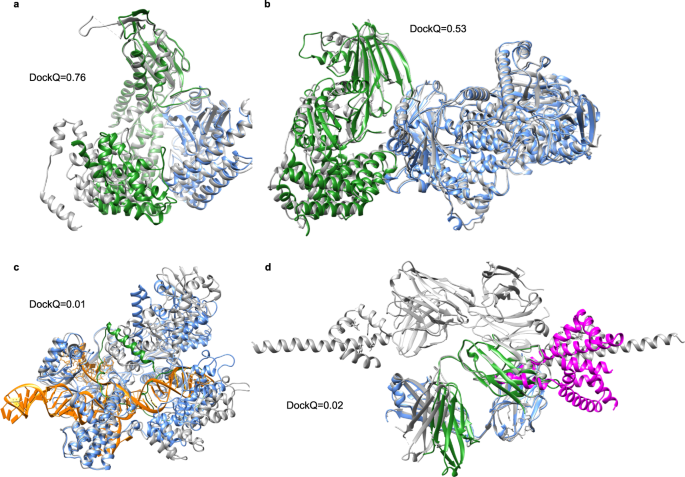
The native structures are represented as grey ribbons. a Docking of 7EIV chains A (blue) and C (green) (DockQ = 0.76). b Docking of 7MEZ chains A (blue) and B (greenish) (DockQ = 0.53). c Prediction of construction 7EL1 chains A (blue) and East (greenish) (DockQ = 0.01). The Dna going through concatenation A is coloured in orangish. d Docking of 7LF7 chains A (blue) and M (magenta) (DockQ = 0.02) and chains B (greenish) and M (magenta) (DockQ = 0.02).
Identifying interacting proteins
Using the best separator from the model ranking, the pDockQ, it is possible to distinguish the 3989 not-interacting proteins from Escherichia coli and the 1481 truly interacting proteins from the test set up with an AUC of 0.87. Another recently published method obtains AUC 0.76 on this set27. However, these results are probably overstated since the negative set only contains bacterial proteins, while the positive set is mainly eukaryotic.
To obtain a more realistic judge, we besides include a set of 1705 not-interacting proteins from mammalian organisms31 combined with the non-interacting proteins from E. coli. On this combined fix of 1481 interacting and 5694 non-interacting proteins, nosotros obtain an AUC of 0.82 for the average interface plDDT and slightly higher (0.84 and 0.85) for the number of interface contacts and residues, respectively (Fig. 5a). pDockQ results in an ROC curve with an AUC of 0.87. Chiefly, pDockQ provides a improve separation at low FPRs, enabling a TPR of 51% at FPR of 1% compared to 27%, 18 and 13% for the interface plDDT, number of interface contacts and residues, respectively. At FPR 5%, the number of interface contacts and residues report TPRs of 49 and 42%, respectively, compared to 43% for the average interface plDDT and 66% for pDockQ. The distribution of the top separators can be seen in Fig. 5b–d.

a The ROC bend as a part of different metrics for discriminating between interacting and non-interacting proteins. IF_plDDT is the average plDDT in the interface, min plDDT per concatenation is the minimum average plDDT of both bondage, average plDDT is the average of the entire complex and IF_contacts and IF_residues are the number of interface residues and contacts respectively. pDockQ is a sigmoidal fit to this with DockQ as the target score, as described above. b–d Distribution of the top discriminating features average interface plDDT (b), the number of interface contacts (c), and d the combination of these (IF_plDDT⋅log(IF_contacts)) and the pDockQ for interacting (non-grey) and non-interacting proteins (grey).
Limitations
Here, nosotros only consider the structures of protein complexes in their heterodimeric state, although each protein chain in these complexes may have homodimer configurations or other higher-order states. It is also possible that the complex itself exists as part of larger biological units, in potentially more complex conformations. Investigating alternative oligomeric states and larger biological assemblies is exterior of the scope of this analysis and left for futurity work.
The study of AF2s ability to divide interacting and non-interacting proteins here contains more extensive data than recent studies27. Still, to exam this separation thoroughly, the data studied hither needs to be extended to compare interactions within individual organisms. We leave this extensive analysis to further studies.
There is a big difference betwixt the functioning of AF2 on the development and exam sets, reporting 39.4% SR vs 57.8% for the AF2 + Paired MSAs. This discrepancy suggests that the performance is highly dependent on the specific interacting partners being predicted. It is not articulate what causes this difference equally the composition in terms of kingdom, found to be very important (Supplementary Fig. 1b) is similar (54% vs 60% Eukaryotic proteins), the MSAs have similar Neff scores (2699 vs. 2764 on average), the proteins are of like sizes (222 vs. 203 AAs on boilerplate), and the number of residues in the interface is similar (139 vs 120 on boilerplate). This leads us to believe that there may be some unknown pick bias in how the sets were chosen. It can be noted that the development is much smaller than the exam set though (216 vs 1481 proteins), which is why performance should be assessed on as large non-redundant datasets as possible.
Findings and future prospects
Here nosotros show that AlphaFold2sixteen (AF2) can predict the structure of many heterodimeric protein complexes, although it is trained to predict the structure of private poly peptide chains. Even using the default settings, it is clear that AF2 is superior to all other tested docking methods, including other Fold and Dock methods17,24, methods based on shape complementarity30,32 and template-based docking. Using optimised MSAs with AF2, we tin can accurately predict the structure of heterodimeric complexes for an unprecedented SR of 62.7% on a large test prepare. The SR is higher in E.coli (76.four%) than in H. sapiens or S. cerevisiae (58.1% and 66.2% respectively).
Further, past analysing the predicted interfaces, nosotros tin predict the DockQ score33 (pDockQ) with an average error of 0.one, resulting in the separation of adequate and incorrect models with an AUC of 0.95. This means that 31% of the models can be called acceptable at a specificity of 99% (or 54% at 90% specificity). Interestingly, no additional constraints are implemented in AF2 to pull two bondage in contact, meaning that chain interactions (and afterwards interface sizes) are exclusively determined past the amount of inter-concatenation signals extracted by the predictor. Assuming that all residues in an interface contribute to the interaction energy could explain why larger interfaces are more than likely to be correctly predicted.
We find that the MSA generation process can be sped upwards substantially at no performance loss (performance increase of i% SR) by merely fusing MSAs from 2 HHblits34 runs on Uniclust3035 instead of using the MSAs from AF2. Fast MSA generation circumvents the principal computational bottleneck in the pipeline. Using pDockQ makes it possible to separate truly interacting from non-interacting proteins with an AUC of 0.87, making information technology possible to identify 51% of interacting proteins at an error rate of one%. The pDockQ score discriminates between both model quality and binary interactions. Therefore, the same pipeline can identify if two proteins interact and the accuracy of their structure.
Never before has the potential for expanding the known structural understanding of protein interactions been this large, at such a modest cost. In that location are currently 64,006 pairwise homo protein interactions in the human reference interactome36. If 31% of these tin be predicted at an fault rate of i%, this results in the construction of xix,842 human heterodimeric protein structures. The computational cost to run all of this is ~5 days on an Nvidia A100 organization and has since the development of the pipeline presented here, deemed FoldDock, been applied37.
Methods
Development set
A set up of heterodimeric complexes from Dockground benchmark four38 is used to develop the pipeline, focusing on the AF2 configuration presented here. This set contains protein pairs, with each chain having at least 50 residues, sharing <30% sequence identity and no crystal packing artefacts. There are 219 protein interactions for which both unbound (single-concatenation) and bound (interacting chains) structures are available. Unbound bondage share at least 97% sequence identity with the bound counterpart and, to facilitate comparisons, not-matching residues are deleted and renumbered to go identical to the unbound analogue. AF2 MSAs could non exist generated for three of the complexes due to retentiveness limitations (1gg2, 2nqd and 2xwb) using a computational node with 128 Gb RAM for the MSA generation and were thus disregarded, resulting in a total of 216 complexes. The dataset consists of 54% Eukaryotic proteins, 38% Bacterial and 8% from mixed kingdoms, e.g., i bacterial protein interacting with one eukaryotic.
Test set
We used 1661 protein complexes with known interfaces from a recent report27 to test the developed pipeline. Here, three large biological assemblies were excluded. These complexes share <30% sequence identity, have a resolution between 1–5 Å and found unique pairs of PFAM domains (no single protein pair take PFAM domains matching that of whatsoever other pair). Some structures failed to be modelled for diverse reasons (run into limitations of data generation), resulting in a total of 1481 structures. These proteins are mainly from H. sapiens (25%), S. cerevisiae (10%), E. coli (five%) and other Eukarya (30%).
107 of the complexes in the test ready lack beta carbons (Cβs), and 50 have overlapping PDB codes with the evolution fix and were therefore excluded. In the MSA generation from AF2, xx MSAs written report MergeMasterSlave errors regarding discrepancies in the number of match states, resulting in a total of 1484 AF2 MSAs. When folding, three of these (5AWF_D-5AWF_B, 2ZXE_B-2ZXE_A and 2ZXE_A-2ZXE_G) study "ValueError: Cannot create a tensor proto whose content is larger than 2GB", leading to a final set of 1481 complexes. DSSP could but be run successfully for 1391 out of the 1481 protein complexes, and we ignored the remainder in the analysis.
For RF, 26 complexes produced out of memory exceptions during prediction using a GPU with 40 Gb RAM and were excluded from the RF analyses, leaving 1455 complexes.
For the mammalian proteins from Negatome, 7 out of 1733 unmarried chains were redundant co-ordinate to Uniprot (C4ZQ83, I0LJR4, I0LL25, K4CRX6, P62988, Q8NI70, Q8T3B2), 34 had no matching species in the MSA pairing, 106 produced out of retention exceptions during prediction using a GPU with 40 Gb RAM, 35 gave a tensor reshape error, and 65 complexes were homodimers, leaving 1715 complexes for this gear up.
CASP14 gear up and novel protein complexes
As an additional test set, we used a set up of vi heterodimers from the CASP14 experiment. In addition, we extracted viii novel protein complexes deposited in PDB afterwards 15 June 2021, which produced no results for at least one chain in each circuitous when submitted to the HHPRED web server (version 01-09-2021)39,40, see Supplementary Table 3. We selected this pocket-sized prepare to test the functioning on information AF2 is guaranteed not to accept seen.
Non-interacting proteins
Two datasets of known non-interacting proteins were used, one from the same report as the positive test set27. Here, all proteins are from E. coli. 2 methods were used to identify non-interacting proteins, kickoff a fix of proteins with no reported interaction signal in Yeast Two-Hybrid Experiments41 and secondly complexes whose individual proteins were found in dissimilar APMS benchmark complexes42. This dataset contains in total 3989 not-interacting pairs.
The second set contains 1964 unique mammalian protein complexes filtered against the IntAct43 dataset from Negatome31. This data deemed "the transmission stringent gear up" contains proteins annotated from the literature with experimental support describing the lack of protein interaction. Some structures in this dataset are homodimers (65) and are therefore excluded, resulting in 1705 structures. Together at that place are 5694 non-interacting poly peptide complexes.
AlphaFold2 default MSA generation methodology
The input to AlphaFold2 (AF2) consists of several MSAs. Nosotros used the AF2 MSA generationxvi, which builds iii different MSAs generated past searching the Large Fantastic Database44 (BFD) with HHBlits34 (from hh-suite v.iii.0-beta.iii version 14/07/2017) and both MGnify v.2018_1245 and Uniref90 5.2020_0146 with jackhmmer from HMMER347. The AF2 MSAs were generated by supplying a concatenated poly peptide sequence of the entire circuitous to the AF2 MSA generating pipeline in FASTA format. The resulting MSAs will thus mainly incorporate gaps for one of the two query proteins in each row, every bit only unmarried bondage can obtain hits in the searched databases (Fig. half-dozen). No trimming or gap removal was performed on these MSAs.
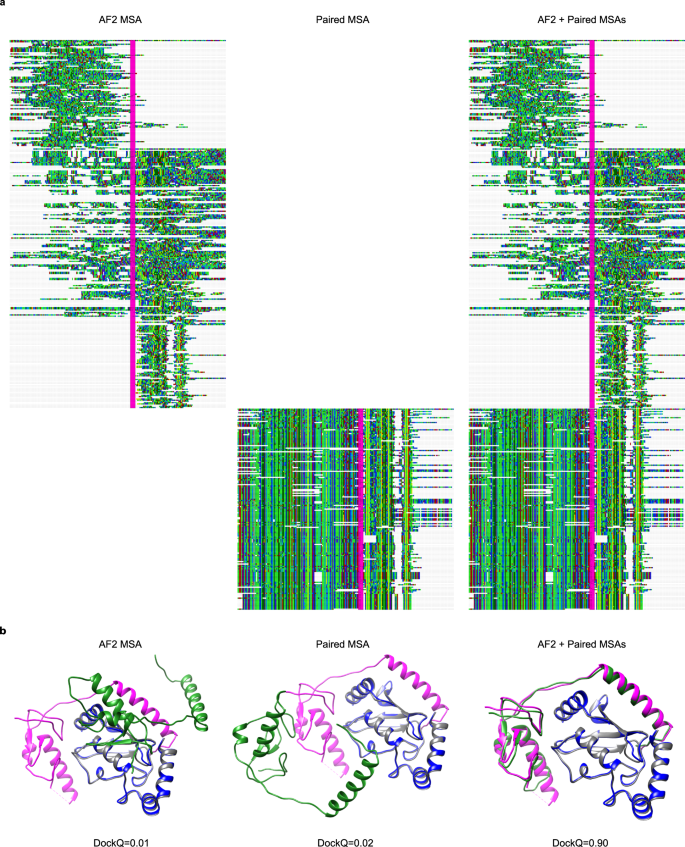
a Delineation of MSAs generated by AF2 and the paired version matched using organism information. Both AF and paired representations are sections containing ten% of the sequences aligned in the original MSA. Concatenated bondage are separated by a vertical line (magenta). The visualisations were fabricated using Jalview version 2.eleven.1.449. b Docking visualisations for PDB ID 5D1M with the model/native chains A in blueish/grey and B in greenish/magenta using the three unlike MSAs in (a). The DockQ scores are 0.01, 0.02 and 0.90 for AF2, paired, and AF2 + paired MSAs, respectively.
MSA block diagonalization
In add-on to the default AF2 MSA, nosotros generated an additional MSA past but concatenating diagonally MSAs generated independently from each of the 2 chains. These MSAs were constructed by running HHblits34 version iii.i.0 against uniclust30_2018_0835 with these options:
hhblits -Due east 0.001 -all -oa3m -n two
The concatenation is done by joining side-by-side the ii input chains; so sequences from one MSA are added, aligned to the corresponding input chain. Each sequence in the MSA is then elongated with gaps (on the right side if it is the left sequence MSA or the other manner around), to reach the length of the two concatenated input chains. The process is and then repeated for the other input chain MSA to complete the block diagonalization.
Paired MSA generation
In add-on to the block diagonalization MSAs, nosotros used a "paired MSA", synthetic using organism information, where sequences are matched based on their organism origins4,21,24 (Fig. half dozen). The rationale behind using a paired MSA is to identify inter-chain co-evolutionary information. An unpaired MSA has a limited inter-chain signal since the chains are treated in isolation.
The organism data was, using the OX identifier, extracted from the ii HHblits MSAs48. Next, all hits with more than xc% gaps were removed. From all remaining hits in the two MSAs, the highest-ranked hit from ane organism was paired with the highest-ranked hit of the interacting chain from the same organism. Pairing the correct sequences should result in MSAs containing inter-chain co-evolutionary information27.
Number of effective sequences (Neff)
To gauge the data in each MSA, we clustered sequences at 62% identity, as described in a previous study50. The number of clusters obtained in this way has been used to indicate a Neff value for each MSA.
Unaligned FASTA sequences were extracted from the three AF2 default MSAs. Obtained sequences were processed with the CD-HIT software51 version 4.vii (http://weizhong-lab.ucsd.edu/cd-hit/) using the options:
-c 0.62 -G 0 -n iii -aS 0.ix
Nosotros calculated the Neff scores separately for paired and AF2 MSAs.
AlphaFold2
Nosotros modelled complexes using AlphaFold2xvi (AF2) by modifying the script https://github.com/deepmind/alphafold/blob/primary/run_alphafold.py to insert a chain break of 200 residues—as suggested in the development of RoseTTAFold17 (RF). During modelling, relaxation was turned off. We note that performing model relaxation did not increase operation in the AF2 paper16 and was, therefore, ignored to save computational toll. No templates were used to build structures, every bit this would not assess the prediction accuracy of unknown structures or structures without sufficient matching templates. Further, AF2 has been shown to perform well for unmarried chains without templates and has reported higher accuracy than template-based methods fifty-fifty when robust templates are available16.
We supplied four different types of MSAs to AF2: (1) the MSAs generated past using the default AF2 settings, (two) the top paired MSAs constructed using HHblits, described above, (iii) both alignments together and finally, (4) the top paired and single-chain MSAs from HHblits to speed upwards predictions (but for the test set). AF2 was run with two different network models, AF2 model_1 (used in CASP14) and AF2 model_1_ptm, for each MSA. The 2d model, model_1_ptm, is a fine-tuned version of model_1 that predicts the TMscore52 and alignment errors16. We ran these two different models past using two different configurations. The configurations utilise a varying amount of recycles and ensemble structures. Recycle refers to the number of times iterative refinement is applied by feeding the intermediate outputs recursively back into the same neural network modules. At each recycling, the MSAs are resampled, assuasive for new information to be passed through the network. The number of ensembles refers to how many times information is passed through the neural network before it is averaged16. The two configurations used are; the CASP14 configuration (3 recycles, eight ensembles) and an increased number of recycles (ten) only just i ensembles.
Since construction prediction with AF2 is a not-deterministic procedure, we generate five models initiated with different seeds. To save computational cost, this was just performed for the best modelling strategy. Nosotros rank the five models for each complex past the number of residues in the interface, giving the best effect.
RoseTTAFold
For comparison, the RoseTTAFold (RF) end-to-end version17 was run using the paired MSAs with the tiptop hits. The RoseTTAFold pipeline for complex modelling only generates MSAs for bacterial protein complexes, while the proteins in our test set are mainly Eukaryotic. Therefore, nosotros utilise the paired alignments here. We compare RF with AF2 using the same inputs (the paired MSAs) for both the development and test datasets to provide a more fair comparing, as AF2 searches many dissimilar databases to obtain as much evolutionary information equally possible when generating its MSAs. To predict the complexes, we use the "concatenation intermission modelling" as suggested in RF (https://github.com/RosettaCommons/RoseTTAFold/tree/master/instance/complex_modeling) using the post-obit command:
predict_complex.py -i msa.a3m -o circuitous -Ls chain1_length chain2_length
No optimisation of the RF protocol was made here.
MDockPP
The docking method MDockPP30 was run through the provided webserver (https://zougrouptoolkit.missouri.edu/MDockPP/). This docking algorithm is based on fast Fourier transform (FFT). The docking results are assessed using the "in-firm" scoring function ITScorePP.
GRAMM
For comparing, a rigid-body docking method, GRAMM32, was used. Here, two poly peptide models are docked using a FFT procedure to generate 340,000 docking poses for each circuitous. The spring structures extracted from complexes in the test set were used as inputs. This docking generation stage mainly considers the geometric surface properties of the two interacting structures, assuasive minor clashes to leave some space for conformational flexibility adjustment. As the bound form of the proteins is used, this should represent an like shooting fish in a barrel case for GRAMM-based docking, and performance drops significantly when unbound structures or models are used53. The atom-atom contact energy AACE18 is used to score and rank all poses, as this has been shown to provide improve results than shape-complementarity solitary54.
Template-based docking
For comparison, a template-based docking protocol7 referred to equally "TMdock" is also adopted. The adopted template library includes 11756 protein complexes obtained from the Dockground database38 (release 28-ten-2020). Monomers from target complexes are structurally aligned with complexes in the supplied libraries (depleted of the target construction PDB ID) in social club to identify the best bachelor template structure. The spring class of the template structures was used. TM scores resulting from the alignment of target proteins to each template are averaged and used to score obtained docking models. Alternatively, nosotros refer to "TMdock Interfaces" when targets are structurally aligned just to the template interfaces, divers as every residue with a Cβ cantlet closer than 12 Å from any Cβ atom in the other chain.
AlphaFold-multimer
The simultaneous fold-and-dock program based on the same principles as AF2, AlphaFold-multimer28, was run with the default settings. These entail creating four different MSAs. Three unlike MSAs are created by searching Uniref90 v.2020_0146, Uniprot 5.2021_0448 and MGnify v.2018_1245 with jackhmmer from HMMER347 and one joint is created by searching the Big Fantastic Database44 (BFD) and uniclust30_2018_0835 with HHBlits34 (from hh-suite v.3.0-beta.3 version 14/07/2017).
The results from the Uniprot search are used for MSA pairing and the results from the remaining searches are used to create a cake-diagonalized MSA, similar to the procedures described above. All four MSAs are then used to fold a protein circuitous. Some complexes failed due to computational limitations, resulting in 1458 out of 1481 complexes successfully folded.
Scoring models
The backbone atoms (Northward, CA and C) were extracted from the predicted AF2 structures (as these are the just predicted atoms in the stop-to-cease version of RF). The interface scoring plan DockQ33 was then run (without whatsoever special settings) to compare the predicted and actual interfaces. This programme compares interfaces using a combination of three dissimilar CAPRI55 quality measures (Fnat, LRMS, and iRMS) converted to a continuous scale, where an acceptable model comprises a DockQ score of at least 0.23.
Ranking models
To analyse the ability of AF2 to distinguish right models every bit well as interacting from non-interacting proteins, we analyse the separation between adequate and incorrect models as a function of different metrics on the development prepare: the number of unique interacting residues (Cβs from different chains within 8 Å from each other), the total number of interactions between Cβs from different bondage (referred to as the number of interface contacts), boilerplate predicted lDDT (plDDT) score from AF2 for the interface, the minimum of the boilerplate plDDT for both chains and the average plDDT over the whole heterodimer.
We use these metrics equally a threshold to build a confusion matrix, where true/false positives (TP and FP respectively) are right/incorrect docking models which places to a higher place the threshold and false/true negatives (FN and TN respectively) are correct/incorrect docking models which scores below the threshold. From the built confusion matrix, we derive the true positive rate (TPR), faux positive charge per unit (FPR) defined as:
$${{{{{\rm{TPR}}}}}}=\frac{{{{{{\rm{TP}}}}}}}{{{{{{\rm{TP}}}}}}+{{{{{\rm{FN}}}}}}}$$
(one)
$${{{{{\rm{FPR}}}}}}=\frac{{{{{{\rm{FP}}}}}}}{{{{{{\rm{FP}}}}}}+{{{{{\rm{TN}}}}}}}$$
(2)
And so, we calculate TPR and FPR for each possible value assumed by the set of dockings given a single metric and plot TPR as a part of FPR in society to obtain an ROC curve. We compute the expanse nether curve (AUC) for ROC curves obtained for each metric to compare different metrics. The AUC is divers as:
$${{{{{\rm{AUC}}}}}}={\int }_{\!\!x=0}^{1}{{{{{\rm{TPR}}}}}}\left(\frac{1}{{{{{{\rm{FPR}}}}}}(x)}\right){{{{{\rm{d}}}}}x}$$
(3)
The TPR and FPR for different thresholds are used to summate the fraction of models that can exist called correct out of all models and the positive predictive value (PPV). The fraction of acceptable and wrong models are obtained past multiplying the TPR and FPR with the SR. Multiplying the FPR with the SR results in the imitation discovery charge per unit (FDR) and the PPV tin can be calculated past dividing the fraction of acceptable models by the sum of the adequate and incorrect models. The PPV, FDR and SR are defined as:
$${{{{{\rm{PPV}}}}}}=\frac{{{{{{\rm{TP}}}}}}}{{{{{{\rm{TP}}}}}}+{{{{{\rm{FP}}}}}}}$$
(4)
$${{{{{\rm{FDR}}}}}}=1-{{{{{\rm{PPV}}}}}}$$
(5)
$${{{{{\rm{SR}}}}}}={{{{{\rm{Fraction}}}}}}\,{{{{{\rm{of}}}}}}\,{{{{{\rm{predicted}}}}}}\,{{{{{\rm{models}}}}}}\,{{{{{\rm{with}}}}}}\,{{{{{\rm{DockQ}}}}}}\ge 0.23$$
(half dozen)
pDockQ
As information technology is not merely desirable to know when a model is accurate only also how accurate this model is, nosotros developed a predicted DockQ score, pDockQ. This score is created by plumbing equipment a sigmoidal curve (Fig. 2c) using "curve_fit" from SciPy 5.1.4.i56, to the DockQ scores using the average interface plDDT multiplied with the logarithm of the number of interface contacts, with the following sigmoidal equation:
$${{{{{\rm{pDockQ}}}}}}=\frac{L}{1+{e}^{-1000(x-{x}_{0})}}+{{{{{\rm{b}}}}}}$$
(7)
where
$$x={{{{{\rm{boilerplate}}}}}}\; {{{{{\rm{interface}}}}}}\; {{{{{\rm{plDDT}}}}}}\cdot {{\log }}({{{{{\rm{number}}}}}}\; {{{{{\rm{of}}}}}}\; {{{{{\rm{interface}}}}}}\; {{{{{\rm{contacts}}}}}})$$
(eight)
and we obtain L = 0.724, 10 0 = 152.611, chiliad = 0.052 and b = 0.018.
Analysis of models
To analyse the possibility of determining when AF2 tin model a circuitous correctly, we analyse the structures and the MSAs. We investigated: the number of constructive sequences (Neff), the secondary structure in the interface annotated using DSSP57, the length of the shortest chain, the number of residues in the interface and the number of contacts in the interface.
DSSP was run on the entire complexes, and the resulting annotations were grouped into 3 categories; helix (3-plow helix (3ten helix), 4-turn helix (α helix) and 5-plough helix (π helix)), sheet (extended strand in parallel or antiparallel β-sheet conformation and residues in isolated β-bridges) and loop (residues which are not in any known conformation).
In addition, we appraise the PPV of the top N interface DCA signals using the paired MSAs. Here, N is the number of true interface contacts (Cβs from dissimilar chains within 8 Å from each other). The PPV is therefore the fraction of the acme Due north DCA signals in the interface that are true contacts. The DCA signals are computed using GaussDCA58.
$${{{{{\rm{Interface}}}}}}\,{{{{{\rm{PPV}}}}}}=\frac{{{{{{\rm{Number}}}}}}\; {{{{{\rm{of}}}}}}\; {{{{{\rm{correct}}}}}}\; {{{{{\rm{contacts}}}}}}\; {{{{{\rm{amid}}}}}}\; {{{{{\rm{tiptop}}}}}}\; {{{{{\rm{N}}}}}}\;{{{{{\rm{interface}}}}}}\; {{{{{\rm{DCA}}}}}}\; {{{{{\rm{signals}}}}}}}{N}$$
(9)
Computational cost
To compare the computation required for each MSA, nosotros compared the time information technology took to generate MSAs for iii protein pairs (PDB: 4G4S_P-O, 5XJL_A-2 and 5XJL_2-M), using either the block diagonalization or AF2 protocol. The tests were performed on a computer using 16 CPU cores from an Intel Xeon E5-2690v4.
Fusing the MSAs took 3 s on average per tested complex. It took 7884 s for generating the AF2 MSAs, the single-chain searches took 338 s on average and the pairing 2 s. The pairing and fusing are thereby negligible compared to searching, resulting in a speedup of 24 times for the hhblits searches. In comparison, folding using the m1-10-1 strategy took 191 s on boilerplate for these pairs.
Reporting summary
Further information on research design is available in the Nature Inquiry Reporting Summary linked to this article.
Data availability
The raw information used in this study, including multiple sequence alignments and predicted PDB files, are available in the figshare from Science for Life laboratory nether accretion lawmaking 16866202.v1. All other data supporting the findings of this report are available within the commodity and its supplementary data files. The results used to produce all figures can be establish in the supplementary information. Additional data and relevant information volition be available from the corresponding author upon reasonable request.
Lawmaking availability
All code to run FoldDock and reproduce the assay here can be obtained here https://gitlab.com/ElofssonLab/FoldDock (commit 2e4c96aa352338976260ece0646ceaaa75392dec) nether the Apache License, Version 2.0.
Change history
-
24 March 2022
A Correction to this paper has been published: https://doi.org/10.1038/s41467-022-29480-five
References
-
Liddington, R. C. Structural Basis of Protein–Protein Interactions. Protein-Poly peptide Interactions 261, three–14 https://doi.org/ten.1385/1-59259-762-ix:003 (2004).
-
Keskin, O., Gursoy, A., Ma, B. & Nussinov, R. Principles of protein-poly peptide interactions: what are the preferred ways for proteins to interact? Chem. Rev. 108, 1225–1244 (2008).
-
Nooren, I. M. A. NEW EMBO Member'Due south REVIEW: diversity of protein-protein interactions. EMBO J. 22, 3486–3492 (2003).
-
Cong, Q., Anishchenko, I., Ovchinnikov, S. & Baker, D. Protein interaction networks revealed by proteome coevolution. Scientific discipline 365, 185–189 (2019).
-
Zhang, Q. C. et al. Structure-based prediction of protein-protein interactions on a genome-broad scale. Nature 490, 556–560 (2012).
-
Marshall, K. R. & Vakser, I. A. Poly peptide-Protein Docking Methods. In Proteomics and Protein-Protein Interactions (ed. Waksman, K.) 115–146 (Springer, 2005).
-
Kundrotas, P. J., Zhu, Z., Janin, J. & Vakser, I. A. Templates are bachelor to model nearly all complexes of structurally characterized proteins. Proc. Natl Acad. Sci. USA 109, 9438–9441 (2012).
-
Porter, Yard. A., Desta, I., Kozakov, D. & Vajda, Due south. What method to use for protein–protein docking? Curr. Opin. Struct. Biol. 55, 1–7 (2019).
-
Halperin, I., Ma, B., Wolfson, H. & Nussinov, R. Principles of docking: An overview of search algorithms and a guide to scoring functions. Proteins 47, 409–443 (2002).
-
Shammas, Due south. L. et al. Insights into Coupled Folding and Binding Mechanisms from Kinetic Studies. J. Biol. Chem, 291, 6689–6695 (2016).
-
Eginton, C., Naganathan, S. & Beckett, D. Sequence-function relationships in folding upon binding. Protein Sci. 24, 200–211 (2015).
-
Andrusier, Due north., Mashiach, East., Nussinov, R. & Wolfson, H. J. Principles of flexible poly peptide-protein docking. Proteins 73, 271–289 (2008).
-
Kurkcuoglu, Z. & Bonvin, A. M. J. J. Pre- and postal service-docking sampling of conformational changes using ClustENM and HADDOCK for protein-protein and protein-DNA systems. Proteins 88, 292–306 (2020).
-
Lensink, M. F. et al. Blind prediction of homo- and hetero-protein complexes: The CASP13-CAPRI experiment. Proteins 87, 1200–1221 (2019).
-
Vreven, T. et al. Updates to the integrated protein-poly peptide interaction benchmarks: docking benchmark version 5 and affinity criterion version 2. J. Mol. Biol. 427, 3031–3041 (2015).
-
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
-
Baek, Chiliad. et al. Accurate prediction of poly peptide structures and interactions using a three-track neural network. Science 373, 871–876 (2021).
-
Kandathil, S. K., Greener, J. G., Lau, A. M. & Jones, D. T. Ultrafast end-to-end protein structure prediction enables loftier-throughput exploration of uncharacterised proteins. Proc. Natl Acad. Sci. The states 119, e2113348119 (2022).
-
Chowdhury, R. et al. Single-sequence protein structure prediction using linguistic communication models from deep learning. Preprint at bioRxiv https://doi.org/x.1101/2021.08.02.454840 (2021).
-
Procaccini, A., Lunt, B., Szurmant, H., Hwa, T. & Weigt, M. Dissecting the specificity of protein-protein interaction in bacterial two-component signaling: orphans and crosstalks. PLoS 1 six, e19729 (2011).
-
Weigt, M., White, R. A., Szurmant, H., Hoch, J. A. & Hwa, T. Identification of direct residue contacts in protein-protein interaction past bulletin passing. Proc. Natl Acad. Sci. USA 106, 67–72 (2009).
-
Hashemifar, S., Neyshabur, B., Khan, A. A. & Xu, J. Predicting protein–poly peptide interactions through sequence-based deep learning. Bioinformatics 34, i802–i810 (2018).
-
Yang, J. et al. Improved protein structure prediction using predicted inter-residue orientations. Preprint at bioRxiv https://doi.org/10.1101/846279 (2019).
-
Pozzati, Thousand. et al. Limits and potential of combined folding and docking using PconsDock. Bioinformatics 38, 954–961 (2021).
-
Lamb, J. & Elofsson, A. pyconsFold: a fast and easy tool for modelling and docking using distance predictions. Bioinformatics https://doi.org/ten.1093/bioinformatics/btab353 (2021).
-
Szurmant, H. & Weigt, Chiliad. Inter-balance, inter-protein and inter-family coevolution: bridging the scales. Curr. Opin. Struct. Biol. 50, 26–32 (2018).
-
Green, A. G. et al. Big-scale discovery of protein interactions at residue resolution using co-evolution calculated from genomic sequences. Nat. Commun. 12, i–12 (2021).
-
Evans, R. et al. Protein complex prediction with AlphaFold-Multimer. Preprint at bioRxiv https://doi.org/10.1101/2021.x.04.463034 (2021).
-
Lensink, Thou. F. et al. Prediction of protein assemblies, the adjacent borderland: The CASP14-CAPRI experiment. Proteins https://doi.org/x.1002/prot.26222 (2021).
-
Huang, South.-Y. & Zou, X. MDockPP: a hierarchical approach for protein-poly peptide docking and its application to CAPRI rounds fifteen-xix. Proteins 78, 3096–3103 (2010).
-
Blohm, P. et al. Negatome 2.0: a database of non-interacting proteins derived by literature mining, transmission annotation and protein construction analysis. Nucleic Acids Res. 42, D396–D400 (2014).
-
Vakser, I. A. Evaluation of GRAMM low-resolution docking methodology on the hemagglutinin-antibody complex. Proteins Suppl 1, 226–230 (1997).
-
Basu, S. & Wallner, B. DockQ: a quality measure for protein-poly peptide docking models. PLoS ONE 11, e0161879 (2016).
-
Steinegger, One thousand. et al. HH-suite3 for fast remote homology detection and deep protein annotation. BMC Bioinforma. twenty, 473 (2019).
-
Mirdita, M. et al. Uniclust databases of clustered and deeply annotated protein sequences and alignments. Nucleic Acids Res. 45, D170–D176 (2017).
-
Luck, K. et al. A reference map of the man binary protein interactome. Nature 580, 402–408 (2020).
-
Shush, D. F. et al. Towards a structurally resolved human poly peptide interaction network. Preprint at bioRxiv https://doi.org/10.1101/2021.11.08.467664 (2021).
-
Kundrotas, P. J. et al. Dockground: a comprehensive information resource for modeling of protein complexes. Protein Sci. 27, 172–181 (2018).
-
Gabler, F. et al. Protein sequence analysis using the MPI bioinformatics toolkit. Curr. Protoc. Bioinforma. 72, e108 (2020).
-
Zimmermann, 50. et al. A completely reimplemented MPI bioinformatics toolkit with a new HHpred server at its core. J. Mol. Biol. 430, 2237–2243 (2018).
-
Rajagopala, South. V. et al. The binary poly peptide-protein interaction landscape of Escherichia coli. Nat. Biotechnol. 32, 285–290 (2014).
-
Kuhlbrandt, W. The resolution revolution. Scientific discipline 343, 1443–1444 (2014).
-
Orchard, S. et al. The MIntAct project-IntAct as a common curation platform for 11 molecular interaction databases. Nucleic Acids Res. 42, D358–D363 (2014).
-
BFD. https://bfd.mmseqs.com/.
-
Mitchell, A. L. et al. MGnify: the microbiome analysis resources in 2020. Nucleic Acids Res. 48, D570–D578 (2020).
-
Suzek, B. E., Huang, H., McGarvey, P., Mazumder, R. & Wu, C. H. UniRef: comprehensive and non-redundant UniProt reference clusters. Bioinformatics 23, 1282–1288 (2007).
-
Eddy, Southward. R. Accelerated Contour HMM Searches. PLoS Comput. Biol. vii, e1002195 (2011).
-
UniProt Consortium. UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res. 49, D480–D489 (2021).
-
Waterhouse, A. M., Procter, J. B., Martin, D. M. A., Clamp, Chiliad. & Barton, One thousand. J. Jalview Version ii-a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191 (2009).
-
Kosciolek, T. & Jones, D. T. Authentic contact predictions using covariation techniques and machine learning. Proteins 84, Suppl one. 145–151 (2016).
-
Li, West., Jaroszewski, 50. & Godzik, A. Clustering of highly homologous sequences to reduce the size of large poly peptide databases. Bioinformatics 17, 282–283 (2001).
-
Zhang, Y. & Skolnick, J. Scoring function for automatic assessment of protein structure template quality. Proteins 57, 702–710 (2004).
-
Singh, A., Dauzhenka, T., Kundrotas, P. J., Sternberg, M. J. East. & Vakser, I. A. Application of docking methodologies to modeled proteins. Proteins 88, 1180–1188 (2020).
-
Anishchenko, I., Kundrotas, P. J. & Vakser, I. A. Contact potential for structure prediction of proteins and poly peptide complexes from Potts model. Biophys. J. 115, 809–821 (2018).
-
Lensink, One thousand. F. & Wodak, Southward. J. Docking and scoring protein interactions: CAPRI 2009. Proteins 78, 3073–3084 (2010).
-
Virtanen, P. et al. SciPy 1.0: cardinal algorithms for scientific computing in Python. Nat. Methods 17, 261–272 (2020).
-
Kabsch, W. & Sander, C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 22, 2577–2637 (1983).
-
Baldassi, C. et al. Fast and accurate multivariate Gaussian modeling of protein families: predicting residual contacts and protein-interaction partners. PLoS 1 9, e92721 (2014).
Acknowledgements
Nosotros thank Petras Kundrotas for supplying the new heterodimeric proteins without templates in the PDB. We also give thanks Liming Qiu and Xiaoqin Zou for their aid with running their docking plan MDockPP in a timely style. Financial support: Swedish Enquiry Council for Natural Science, grant No. VR-2016-06301 and Swedish Eastward-scientific discipline Research Center. Computational resources: Swedish National Infrastructure for Computing, grants: SNIC 2021/5-297, SNIC 2021/6-197 and Berzelius-2021-29. All fiscal back up and computational resources were received by A.E.
Funding
Open admission funding provided by Stockholm University.
Author data
Affiliations
Contributions
P.B. and G.P. performed the studies; all authors contributed to the assay. P.B. wrote the kickoff typhoon of the manuscript; all authors contributed to the last version. A.E. obtained funding.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Rodrigo Honorato and Shoshana Wodak for their contribution to the peer review of this work. Peer reviewer reports are available.
Additional information
Publisher'due south notation Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, every bit long equally yous give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party cloth in this commodity are included in the article'south Creative Commons license, unless indicated otherwise in a credit line to the cloth. If textile is non included in the article'southward Creative Commons license and your intended use is non permitted by statutory regulation or exceeds the permitted use, you will demand to obtain permission direct from the copyright holder. To view a re-create of this license, visit http://creativecommons.org/licenses/by/4.0/.
Reprints and Permissions
About this article
Cite this article
Bryant, P., Pozzati, G. & Elofsson, A. Improved prediction of protein-protein interactions using AlphaFold2. Nat Commun 13, 1265 (2022). https://doi.org/10.1038/s41467-022-28865-w
-
Received:
-
Accepted:
-
Published:
-
DOI : https://doi.org/10.1038/s41467-022-28865-w
Comments
By submitting a annotate you agree to abide by our Terms and Community Guidelines. If you observe something abusive or that does not comply with our terms or guidelines delight flag it every bit inappropriate.
Source: https://www.nature.com/articles/s41467-022-28865-w
0 Response to "what to do once you discover a binding partner protein"
Post a Comment